Max W. Shen

Hi, I am a Principal Scientist at Genentech, Prescient Frontiers Research. I research machine learning and biology. Forbes 30 Under 30 Science.
 maxwshen [at] gmail.com
maxwshen [at] gmail.com
 Google Scholar
Google Scholar
 SF Bay Area, CA, USA
SF Bay Area, CA, USA
I've led research published in Nature, Cell (with a front cover featuring my pixel artwork), Nature Biotechnology, ICML, and invited for talks at NeurIPS workshops on ML for biology. I did my PhD at the Broad Institute and MIT's Computer Science & Artificial Intelligence Laboratory with David R. Liu, Aviv Regev, and formerly, David Gifford. Before that, I graduated summa cum laude from U.C. San Diego.
Service: Area chair and co-organizer of ICLR Workshop on Machine Learning for Drug Discovery, 2022 & 2023.
Publications
Towards Understanding and Improving GFlowNet Training
Max W Shen, Emmanuel Bengio, Ehsan Hajiramezanali, Andreas Loukas, Kyunghyun Cho, Tommaso Biancalani
Accepted at ICML 2023
Deep Fitness Inference for Drug Discovery with Directed Evolution
Nathaniel Diamant, Christina Helmling, Kangway Chuang, Christian Cunningham, Tommaso Biancalani, Gabriele Scalia*, Max W Shen*
Invited talk at NeurIPS 2022, Learning Meaningful Representations of Life Workshop
Reconstruction of evolving gene variants and fitness from short sequencing reads
Max W Shen, Kevin T. Zhao, David R. Liu
Invited talk at NeurIPS 2020, Learning Meaningful Representations of Life Workshop
Nature Chemical Biology, 2021.

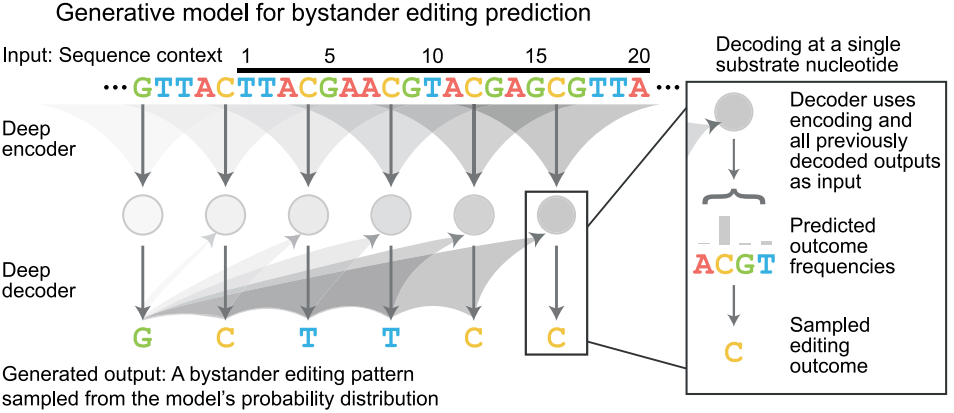
Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning
Max W Shen*, Mandana Arbab*, Beverly Mok, Christopher Wilson, Żaneta Matuszek, Christopher A. Cassa, David R. Liu
Cell, 2020. My artwork was featured on the cover of the July 23, 2020 issue!
[Co-first author reordering approved by all co-first authors]
[Code] [Interactive web app]
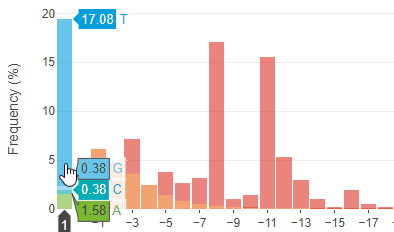
Predictable and precise template-free CRISPR editing of pathogenic variants
Max W Shen*, Mandana Arbab*, Jonathan Y Hsu, Daniel Worstell, Sannie J Culbertson, Olga Krabbe, Christopher A Cassa, David R Liu, David K Gifford, Richard I Sherwood
Nature, 2018
[Code] [Interactive web app] [Press feature by Dash plotly for data visualization]
Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis and machine learning
Luke W Koblan*, Max W Shen*, Mandana Arbab*, Jeffrey A Hussmann, Andrew V Anzalone, Jordan L Doman, Gregory A Newby, Dian Yang, Beverly Mok, Joseph M Replogle, Albert Xu, Tyler A Sisley, Jonathan S Weissman, Britt Adamson, David R Liu
[Co-first author reordering approved by all co-first authors]
Nature Biotechnology, 2021.
Small molecule inhibition of ATM kinase increases CRISPR-Cas9 1-bp insertion frequency
Heysol C Bermudez-Cabrera, Sannie Culbertson, Sammy Barkal, Benjamin Holmes, Max W Shen, Sophia Zhang, David K Gifford, Richard I Sherwood
Nature Communications, 2021.
Detection of gene cis-regulatory element perturbations in single cell transcriptomes
Grace H.T. Yeo, Oscar Juez, Qing Chen, Budhaditya Banerjee, Lendy Chu, Max W Shen, May Sabry, Ive Logister, Richard I Sherwood, David K Gifford
PLOS Computational Biology, 2021.
PrimeDesign software for rapid and simplified design of prime editing guide RNAs
Jonathan Y Hsu, Julian Grünewald, Regan Szalay, Justine Shih, Andrew V Anzalone, Kin Chung Lam, Max W Shen, Karl Petri, David R Liu, Keith Joung, Luca Pinello
Nature Communications, 2021.
Machine learning based CRISPR gRNA design for therapeutic exon skipping
Wilson Louie, Max W Shen, Zakir Tahiry, Sophia Zhang, Daniel Worstell, Christopher A Cassa, Richard I Sherwood
PLOS Computational Biology, 2021.
Comprehensive Mapping of Key Regulatory Networks that Drive Oncogene Expression
Lin Lin, Benjamin Holmes, Max W. Shen, Darnell Kammeron, Niels Geijsen, David K Giffiord, Richard I Sherwood
Cell Reports, 2020.
Continuous evolution of SpCas9 variants compatible with non-G PAMs
Shannon M Miller*, Tina Wang*, Peyton B Randolph, Mandana Arbab, Max W Shen, Tony P Huang, Zaneta Matuszek, Gregory A Newby, Holly A Rees, David R Liu
Nature Biotechnology, 2020
[Code]
Assembly of long error-prone reads using de Bruijn graphs
Yu Lin*, Jeffrey Yuan*, Mikhail Kolmogorov, Max W Shen, Mark Chaisson, Pavel A Pevzner
Proceedings of the National Academy of Sciences, 2016
plasmidSPAdes: assembling plasmids from whole genome sequencing data
Dmitry Antipov, Nolan Hartwick, Max W Shen, Mikahil Raiko, Alla Lapidus, Pavel A. Pevzner
Bioinformatics, 2016
MEG source imaging method using fast L1 minimum-norm and its applications to signals with brain noise and human resting-state source amplitude images
Ming-Xiong Huang, Charles W Huang, Ashley Robb, AnneMarie Angeles, Sharon L Nichols, Dewleen G Baker, Tao Song, Deborah L Harrington, Rebecca J Theilmann, Ramesh Srinivasan, David Heister, Mithun Diwakar, Jose M Canive, J Christopher Edgar, Yu-Han Chen, Zhengwei Ji, Max W Shen, Fady El-Gabalawy, Michael Levy, Robert McLay, Jennifer Webb-Murphy, Thomas T Liu, Angela Drake, Roland R Lee
Neuroimage, 2014
Technical writings
Hierarchically branched diffusion models for efficient and interpretable multi-class conditional generation
Alex Tseng, Tommaso Biancalani, Max W Shen, Gabriele Scalia
2023
Conditional Diffusion with Less Explicit Guidance via Model Predictive Control
Max W Shen, Ehsan Hajiramezanali, Gabriele Scalia, Alex Tseng, Nathaniel Diamant, Tommaso Biancalani
arXiv preprint, 2022
Trust in AI: Interpretability is not necessary or sufficient, while black-box interaction is necessary and sufficient
Max W Shen
arXiv preprint, 2022.
Stochastic gradient descent implicitly regularizes towards invariant models
Max W Shen, Anastasiya Belyaeva
2021.
Teaching
Causal Inference & Deep Learning
MIT independent activites period, Jan. 2018
Max W. Shen, Fredrik Johansson
○ Prepared and co-taught a short graduate-level class with 4 sessions and 6 total h. Typical attendance: 20 students.
Applied Probabilistic Programming & Bayesian Machine Learning
MIT independent activites period, Jan. 2017
Max W. Shen, Alvin Shi, Carles Boix
○ Prepared and co-taught a short upper-division class with 6 sessions and 9 total h. First class attendance: 100 students, typical attendance: 25 students.
Max W. Shen is a Principal Scientist at Genentech.
This website was designed with Skeleton CSS.
 CV
CV  GitHub
GitHub Twitter
Twitter